category_news
Population genetics of reproductive dormancy in Drosophila
After improving our understanding of the physiological basis of reproductive dormancy in Drosophila, we proceeded to the genetic analysis of the trait. We used (Pool-GWAS) to identify the genetic variants contributing to dormancy in a natural Drosophila population. We have already identified some promising gene candidates, which we will validate through a functional analysis.
ESR update - Dormancy is a well-studied adaptation to facilitate overwintering. Our goal is the identification and characterization of naturally occurring genetic variation affecting reproductive dormancy in Drosophila. Earlier experiments of ours showed unambiguous evidence and support for an alternative classification of the dormancy phenotypes. They further indicated that dormancy may not differ from other responses, corroborating the theory of general stress response against diverse stressful conditions in Drosophila. Finally, dormancy may have a more ancient ancestry than previously believed and may not have been the presumed determinative adaptation that led to the expansion of the Drosophila distribution. We recently published our findings regarding the physiology and evolution of dormancy in Drosophila (Lirakis et al. 2018).
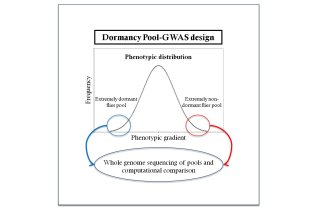
Having understood dormancy in Drosophila in the physiological and evolutionary level allowed us to proceed to the genetic analysis of the trait with more confidence. We planned to perform a genome-wide association study coupled with pooled sequencing (Pool-GWAS) (Schlötterer et al. 2014), a genotype-phenotype mapping scheme in which extreme individuals across a phenotypic gradient are grouped and sequenced as pools. By comparing the allele frequencies between the pools, one can identify the causative genetic variants that explain the phenotypic differences. It is an alternative to individual sequencing and thus a cost-effective method for investigating a large sample of natural variation.
Our earlier experiment showed significant dormancy differences within and between species in Drosophila. However, at very low temperatures (8°C) dormancy response was particularly strong and phenotypic variation was reduced. Thus, we restricted our genetic analysis between 10°C and 12°C dormancy-inducing conditions. We screened a single Drosophila population (comprised of more than 1000 isofemale lines) for dormancy at both temperatures. Such a genetic design allowed us to achieve homogeneous genetic background and satisfying discrimination between opposite phenotypes. We recently obtained the sequencing data, after whole-genome sequencing of pools of individuals with opposite dormancy phenotypes. The computational analysis is in progress, but we have already identified some promising gene candidates. We will further validate these candidates through a functional analysis.
References
Lirakis, M., Dolezal, M. & Schlötterer, C., 2018. Redefining reproductive dormancy in Drosophila as a general stress response to cold temperatures. Journal of Insect Physiology, 107, pp.175–185.
Schlötterer, C. et al., 2014. Sequencing pools of individuals — mining genome-wide polymorphism data without big funding. Nature Reviews Genetics, 15(11), pp.749–763. Available at: http://dx.doi.org/10.1038/nrg3803.